DC Science Series #2 - Rafael Montenegro Marín (DC11)
Welcome to the SYNSENSO DC Science Series. In this series of blog posts, we talk about the scientific projects we are working on as Doctoral Candidates in the SYNSENSO network and share what we are discovering through our research. Enjoy!
Hey!
I am Rafael Montenegro Marín, DC11, within the SYNSENSO network. I am from Costa Rica but have been in Europe for several years. I majored in Biotechnology Engineering back in Costa Rica, where I was inspired to study engineering biology. This motivation came particularly from participating in the iGEM 2016 competition. In addition, I worked for two years doing research in Environmental Microbiology before resuming my studies focused on synthetic biology.
Deciding to explore this field further, I moved to Paris to study for my master’s in Systems and Synthetic Biology. During my master’s internship, I had the opportunity to build a molecular tool to predict protein expression cost by estimating resource competition. With this tool, we can add different sequences with varying codon compositions to predict their impact on fluorescent protein expression. The reduction in fluorescence indicates the cost of expressing a particular sequence. The final goal of that project is to develop tools that enable better designs to express biological circuits in cells.
Motivated to continue exploring the field of synthetic biology, I joined the SYNSENSO network to develop tools for biosensing.
Overview: Intein-based logic gates for multiplexed designs
My project within the SYNSENSO network broadly aims to use split intein proteins to build novel logic gates that could improve biosensing in cell-free systems. Split inteins are proteins capable of ligating two proteins at the peptide level while simultaneously excising themselves from the fused proteins in a reaction called protein trans-splicing (PTS; details of the reaction in the middle panel of Fig. 1)¹. Based on this ligation reaction, split intein proteins have been applied as logic gates for biotechnological applications such as biosensing and biocomputation².
Logic gates are concepts borrowed from electronic circuits to describe biological systems. A logic gate generally refers to a system that receives inputs and triggers output signals. In a biological system, this logic can be interpreted through the following sequential steps: (i) the input signal refers to a target molecule being detected; (ii) next, an operation occurs that corresponds to an enzymatic reaction; and (iii) finally, this enzymatic reaction gives rise to an output signal. Split inteins are ideal triggers for releasing an output signal due to their PTS reaction. Therefore, split inteins are strong candidates for creating novel logic gate designs, especially considering their advantages: fast-reacting, modular, and particularly, their logic functionality¹,².
The main idea and novelty of the project lie in designing and characterising a chimeric protein containing split inteins to perform logic operations (Fig. 1). A chimeric protein is the fusion of proteins or their domains to either enhance a function or introduce a novel one. In theory, our chimeric protein design will detect nucleic acids via recognition domains and, consequently, release an output signal through the action of the split inteins. We are currently designing and screening such a chimeric construct by implementing a directed evolution pipeline. Ultimately, our project thus aims to create a foundational biosensing technology using the chimeric protein design and to demonstrate its biocomputational potential in cell-free biosensing for nucleic acid targets, mainly RNA.
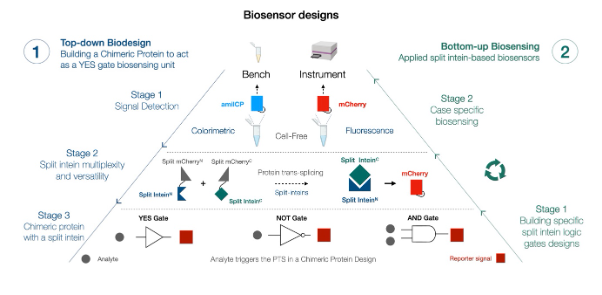
Figure 1: A 2-step biosensor design approach: top-down biodesign for bottom-up biosensing.
The technology for biosensing consists of a two-step approach. The first step is to (i) build a chimeric protein that recognises an analyte to release an mCherry signal through the action of a split intein in a reaction called protein trans-splicing (PTS). This reaction reconstitutes the mCherry molecule and releases it from the system to be detected. The second step is to (ii) couple the detection of a single or multiple analytes, following a defined logic, to biosense a particular condition or disease by tracking a reporter molecule such as mCherry or amilCP.
References
[1] Shah, N. H. & Muir, T. W. Inteins: nature’s gift to protein chemists. Chem. Sci. 16 (2014).
[2] Pinto, F., Thornton, E. L. & Wang, B. An expanded library of orthogonal split inteins enables modular multi-peptide assemblies. Nat. Commun.11, 1529 (2020).

Text by Rafael Montenegro Marín, DC 11 (Intein-based logic gates for multiplexed designs). To find out more about Rafael, visit his profile.
